| Home | Notebook | Data Table | Poster |
Project Code
Goals:
- to predict heart/stroke problems, they are indicated as variables LAHCA7 and LAHCA8 respectively.
- to identify some variables you think would be relevant and trying out some summary using pandas, for example, get the percentage of categorical variables(ethnicity, marital status), average for continuous variables(age, income, etc), or plotting
Import packages and code
import pandas as pd
import numpy as np
import IPython
import matplotlib.pyplot as plt
from sklearn import preprocessing
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
import seaborn as sns
sns.set(style="white")
sns.set(style="whitegrid", color_codes=True)
/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.
import pandas.util.testing as tm
persons_data = pd.read_csv('https://raw.githubusercontent.com/MaxShalom/arise/master/personsx1.csv', delimiter = ',')
persons_data.head()
| FPX | AGE_CHG | INTV_QRT | SEX | NOWAF | FSPOUS2 | COHAB1 | COHAB2 | FCOHAB3 | ASTATFLG | CSTATFLG | FMX | RRP | FRRP | ORIGIN_I | HISPAN_I | RACRECI3 | QCADULT | QCCHILD | R_MARITL | MRACRPI2 | RACERPI2 | HISCODI3 | MRACBPI2 | AGE_P | HHREFLG | RECTYPE | SRVY_YR | FMREFLG | FMRPFLG | PARENTS | DAD_DEGP | MOM_DEGP | SIB_DEGP | CDCMSTAT | DAD_ED | MOM_ED | FMOTHER1 | FFATHER1 | HHX | ... | MAFLG | CHFLG | OPFLG | OGFLG | WHONAM1 | WHONAM2 | NOTCOV | PRPLPLUS | PWRKBR1 | COVER | COVER65O | COVER65 | REGIONBR | WHYNOWKP | GEOBRTH | YRSINUS | CITIZENP | DOINGLWP | WRKLYR1 | WRKHRS2 | PLBORN | HEADST | HEADSTV1 | ARMFVER | ARMFEV | ARMFFC | VACOV | WRKFTALL | WRKMYR | HIEMPOF | EDUC1 | ERNYR_P | ARMFTM7P | ARMFTM1P | ARMFTM2P | ARMFTM3P | ARMFTM4P | ARMFTM5P | ARMFTM6P | ENGLANG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | NaN | 1 | 1 | NaN | NaN | NaN | NaN | NaN | 1 | NaN | 1 | 1 | 1 | 2 | 12 | 3 | NaN | NaN | 4 | 11 | 4 | 4 | 7 | 75 | P | 20 | 2018 | P | B | 4 | NaN | NaN | NaN | 5 | NaN | NaN | 0 | 0 | 9 | ... | NaN | NaN | NaN | NaN | 1.0 | NaN | 2 | 2.0 | NaN | NaN | 1.0 | 1.0 | 1 | 3.0 | 1 | NaN | 1 | 5 | 2 | NaN | 1 | NaN | NaN | NaN | 2 | NaN | NaN | NaN | NaN | NaN | 10 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1 |
| 1 | 1 | NaN | 1 | 1 | NaN | 2.0 | NaN | NaN | NaN | 1 | NaN | 1 | 1 | 1 | 2 | 12 | 1 | NaN | NaN | 1 | 1 | 1 | 2 | 1 | 68 | P | 20 | 2018 | P | B | 4 | NaN | NaN | NaN | 3 | NaN | NaN | 0 | 0 | 20 | ... | NaN | NaN | NaN | NaN | 1.0 | NaN | 2 | 2.0 | NaN | NaN | 1.0 | 1.0 | 1 | 3.0 | 1 | NaN | 1 | 5 | 2 | NaN | 1 | NaN | NaN | NaN | 2 | NaN | NaN | NaN | NaN | NaN | 16 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1 |
| 2 | 1 | NaN | 1 | 1 | NaN | NaN | NaN | NaN | NaN | 1 | NaN | 1 | 1 | 1 | 2 | 12 | 2 | NaN | NaN | 4 | 2 | 2 | 3 | 2 | 76 | P | 20 | 2018 | P | B | 4 | NaN | NaN | NaN | 5 | NaN | NaN | 0 | 0 | 21 | ... | NaN | NaN | NaN | NaN | NaN | NaN | 2 | NaN | NaN | NaN | 2.0 | 2.0 | 1 | 3.0 | 1 | NaN | 1 | 5 | 2 | NaN | 1 | NaN | NaN | NaN | 2 | NaN | NaN | NaN | NaN | NaN | 14 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1 |
| 3 | 1 | NaN | 1 | 2 | NaN | NaN | NaN | NaN | NaN | 1 | NaN | 1 | 1 | 1 | 2 | 12 | 1 | NaN | NaN | 4 | 1 | 1 | 2 | 1 | 67 | P | 20 | 2018 | P | B | 4 | NaN | NaN | NaN | 5 | NaN | NaN | 0 | 0 | 22 | ... | NaN | NaN | NaN | NaN | 1.0 | NaN | 2 | 2.0 | NaN | NaN | 1.0 | 1.0 | 1 | 9.0 | 1 | NaN | 1 | 5 | 2 | NaN | 1 | NaN | NaN | NaN | 2 | NaN | NaN | NaN | NaN | NaN | 14 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1 |
| 4 | 1 | NaN | 1 | 1 | NaN | NaN | NaN | NaN | NaN | 1 | NaN | 1 | 1 | 1 | 2 | 12 | 1 | NaN | NaN | 5 | 1 | 1 | 2 | 1 | 65 | P | 20 | 2018 | P | B | 4 | NaN | NaN | NaN | 2 | NaN | NaN | 0 | 0 | 24 | ... | NaN | NaN | NaN | NaN | NaN | NaN | 2 | NaN | NaN | NaN | 4.0 | 5.0 | 1 | NaN | 1 | NaN | 1 | 1 | 1 | 40.0 | 1 | NaN | NaN | NaN | 1 | 1.0 | NaN | NaN | 4.0 | 2.0 | 14 | 2.0 | NaN | 2.0 | 2.0 | 1.0 | 1.0 | NaN | NaN | 1 |
5 rows × 602 columns
Sex count
1 = Male, 2 = Female
persons_data.groupby("SEX").count()
| FPX | AGE_CHG | INTV_QRT | NOWAF | FSPOUS2 | COHAB1 | COHAB2 | FCOHAB3 | ASTATFLG | CSTATFLG | FMX | RRP | FRRP | ORIGIN_I | HISPAN_I | RACRECI3 | QCADULT | QCCHILD | R_MARITL | MRACRPI2 | RACERPI2 | HISCODI3 | MRACBPI2 | AGE_P | HHREFLG | RECTYPE | SRVY_YR | FMREFLG | FMRPFLG | PARENTS | DAD_DEGP | MOM_DEGP | SIB_DEGP | CDCMSTAT | DAD_ED | MOM_ED | FMOTHER1 | FFATHER1 | HHX | WTIA | ... | MAFLG | CHFLG | OPFLG | OGFLG | WHONAM1 | WHONAM2 | NOTCOV | PRPLPLUS | PWRKBR1 | COVER | COVER65O | COVER65 | REGIONBR | WHYNOWKP | GEOBRTH | YRSINUS | CITIZENP | DOINGLWP | WRKLYR1 | WRKHRS2 | PLBORN | HEADST | HEADSTV1 | ARMFVER | ARMFEV | ARMFFC | VACOV | WRKFTALL | WRKMYR | HIEMPOF | EDUC1 | ERNYR_P | ARMFTM7P | ARMFTM1P | ARMFTM2P | ARMFTM3P | ARMFTM4P | ARMFTM5P | ARMFTM6P | ENGLANG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEX | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 4208 | 0 | 4208 | 2356 | 2020 | 230 | 145 | 230 | 4208 | 0 | 4208 | 4208 | 4208 | 4208 | 4208 | 4208 | 2 | 0 | 4208 | 4208 | 4208 | 4208 | 4208 | 4208 | 2542 | 4208 | 4208 | 2578 | 2431 | 4208 | 279 | 484 | 55 | 4208 | 0 | 0 | 4208 | 4208 | 4208 | 4208 | ... | 6 | 0 | 0 | 0 | 1652 | 63 | 4208 | 1635 | 203 | 2356 | 1852 | 1852 | 4208 | 3225 | 4208 | 449 | 4208 | 4208 | 4208 | 975 | 4208 | 0 | 0 | 12 | 4208 | 1305 | 924 | 380 | 1143 | 918 | 4208 | 1143 | 216 | 1039 | 1234 | 1216 | 1035 | 698 | 368 | 4208 |
| 2 | 5080 | 0 | 5080 | 2581 | 1841 | 243 | 154 | 243 | 5080 | 0 | 5080 | 5080 | 5080 | 5080 | 5080 | 5080 | 6 | 0 | 5080 | 5080 | 5080 | 5080 | 5080 | 5080 | 3248 | 5080 | 5080 | 3286 | 3442 | 5080 | 152 | 309 | 76 | 5080 | 0 | 0 | 5080 | 5080 | 5080 | 5080 | ... | 14 | 0 | 4 | 0 | 2086 | 75 | 5080 | 2065 | 203 | 2581 | 2499 | 2499 | 5080 | 4161 | 5080 | 669 | 5080 | 5080 | 5080 | 962 | 5080 | 0 | 0 | 3 | 5080 | 99 | 75 | 452 | 1095 | 880 | 5080 | 1095 | 7 | 90 | 84 | 80 | 47 | 22 | 11 | 5080 |
2 rows × 601 columns
Value count of disease types
LAHCA7: Persons 18+ years who have at least one limitation due to a heart problem
Heart problem causes limitation:
1 - Mentioned
2 - Not mentioned
7 - Refused
8 - Not ascertained
9 - Don’t know
persons_data["LAHCA7"].unique()
persons_data["LAHCA7"].value_counts()
sns.countplot(x="LAHCA7",data=persons_data,palette="hls")
plt.show()
persons_data_sub=persons_data.loc[persons_data['LAHCA7'] <=2]
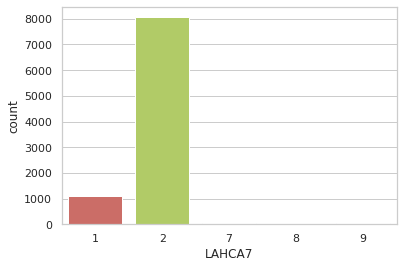
persons_data.groupby("LAHCA7").count()
| FPX | AGE_CHG | INTV_QRT | SEX | NOWAF | FSPOUS2 | COHAB1 | COHAB2 | FCOHAB3 | ASTATFLG | CSTATFLG | FMX | RRP | FRRP | ORIGIN_I | HISPAN_I | RACRECI3 | QCADULT | QCCHILD | R_MARITL | MRACRPI2 | RACERPI2 | HISCODI3 | MRACBPI2 | AGE_P | HHREFLG | RECTYPE | SRVY_YR | FMREFLG | FMRPFLG | PARENTS | DAD_DEGP | MOM_DEGP | SIB_DEGP | CDCMSTAT | DAD_ED | MOM_ED | FMOTHER1 | FFATHER1 | HHX | ... | MAFLG | CHFLG | OPFLG | OGFLG | WHONAM1 | WHONAM2 | NOTCOV | PRPLPLUS | PWRKBR1 | COVER | COVER65O | COVER65 | REGIONBR | WHYNOWKP | GEOBRTH | YRSINUS | CITIZENP | DOINGLWP | WRKLYR1 | WRKHRS2 | PLBORN | HEADST | HEADSTV1 | ARMFVER | ARMFEV | ARMFFC | VACOV | WRKFTALL | WRKMYR | HIEMPOF | EDUC1 | ERNYR_P | ARMFTM7P | ARMFTM1P | ARMFTM2P | ARMFTM3P | ARMFTM4P | ARMFTM5P | ARMFTM6P | ENGLANG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAHCA7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 1103 | 0 | 1103 | 1103 | 414 | 456 | 46 | 36 | 46 | 1103 | 0 | 1103 | 1103 | 1103 | 1103 | 1103 | 1103 | 0 | 0 | 1103 | 1103 | 1103 | 1103 | 1103 | 1103 | 722 | 1103 | 1103 | 727 | 700 | 1103 | 28 | 50 | 8 | 1103 | 0 | 0 | 1103 | 1103 | 1103 | ... | 2 | 0 | 1 | 0 | 412 | 17 | 1103 | 405 | 33 | 414 | 689 | 689 | 1103 | 978 | 1103 | 121 | 1103 | 1103 | 1103 | 133 | 1103 | 0 | 0 | 1 | 1103 | 236 | 168 | 69 | 163 | 120 | 1103 | 163 | 40 | 187 | 230 | 235 | 208 | 146 | 77 | 1103 |
| 2 | 8062 | 0 | 8062 | 8062 | 4453 | 3351 | 415 | 259 | 415 | 8062 | 0 | 8062 | 8062 | 8062 | 8062 | 8062 | 8062 | 6 | 0 | 8062 | 8062 | 8062 | 8062 | 8062 | 8062 | 4998 | 8062 | 8062 | 5065 | 5101 | 8062 | 400 | 734 | 118 | 8062 | 0 | 0 | 8062 | 8062 | 8062 | ... | 18 | 0 | 3 | 0 | 3275 | 119 | 8062 | 3244 | 371 | 4453 | 3609 | 3609 | 8062 | 6323 | 8062 | 982 | 8062 | 8062 | 8062 | 1767 | 8062 | 0 | 0 | 13 | 8062 | 1149 | 814 | 746 | 2037 | 1642 | 8062 | 2037 | 179 | 927 | 1071 | 1042 | 862 | 565 | 297 | 8062 |
| 7 | 53 | 0 | 53 | 53 | 32 | 25 | 4 | 1 | 4 | 53 | 0 | 53 | 53 | 53 | 53 | 53 | 53 | 2 | 0 | 53 | 53 | 53 | 53 | 53 | 53 | 34 | 53 | 53 | 35 | 32 | 53 | 1 | 3 | 2 | 53 | 0 | 0 | 53 | 53 | 53 | ... | 0 | 0 | 0 | 0 | 26 | 1 | 53 | 26 | 0 | 32 | 21 | 21 | 53 | 37 | 53 | 2 | 53 | 53 | 53 | 15 | 53 | 0 | 0 | 0 | 53 | 8 | 7 | 5 | 16 | 15 | 53 | 16 | 2 | 6 | 7 | 8 | 5 | 4 | 2 | 53 |
| 8 | 17 | 0 | 17 | 17 | 12 | 9 | 1 | 0 | 1 | 17 | 0 | 17 | 17 | 17 | 17 | 17 | 17 | 0 | 0 | 17 | 17 | 17 | 17 | 17 | 17 | 11 | 17 | 17 | 11 | 12 | 17 | 1 | 3 | 0 | 17 | 0 | 0 | 17 | 17 | 17 | ... | 0 | 0 | 0 | 0 | 10 | 1 | 17 | 10 | 0 | 12 | 5 | 5 | 17 | 8 | 17 | 1 | 17 | 17 | 17 | 9 | 17 | 0 | 0 | 0 | 17 | 2 | 2 | 3 | 9 | 9 | 17 | 9 | 0 | 2 | 2 | 2 | 1 | 0 | 0 | 17 |
| 9 | 53 | 0 | 53 | 53 | 26 | 20 | 7 | 3 | 7 | 53 | 0 | 53 | 53 | 53 | 53 | 53 | 53 | 0 | 0 | 53 | 53 | 53 | 53 | 53 | 53 | 25 | 53 | 53 | 26 | 28 | 53 | 1 | 3 | 3 | 53 | 0 | 0 | 53 | 53 | 53 | ... | 0 | 0 | 0 | 0 | 15 | 0 | 53 | 15 | 2 | 26 | 27 | 27 | 53 | 40 | 53 | 12 | 53 | 53 | 53 | 13 | 53 | 0 | 0 | 1 | 53 | 9 | 8 | 9 | 13 | 12 | 53 | 13 | 2 | 7 | 8 | 9 | 6 | 5 | 3 | 53 |
5 rows × 601 columns
Data.unique
persons_data_sub.loc[persons_data_sub['LAHCA7'] == 2, 'LAHCA7'] = 0
persons_data_sub["LAHCA7"].unique()
/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:966: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
self.obj[item] = s
array([1, 0])
Data groups (mean, describe)
persons_data_sub.groupby('LAHCA7').mean()
| FPX | AGE_CHG | INTV_QRT | SEX | NOWAF | FSPOUS2 | COHAB1 | COHAB2 | FCOHAB3 | ASTATFLG | CSTATFLG | FMX | RRP | FRRP | ORIGIN_I | HISPAN_I | RACRECI3 | QCADULT | QCCHILD | R_MARITL | MRACRPI2 | RACERPI2 | HISCODI3 | MRACBPI2 | AGE_P | RECTYPE | SRVY_YR | PARENTS | DAD_DEGP | MOM_DEGP | SIB_DEGP | CDCMSTAT | DAD_ED | MOM_ED | FMOTHER1 | FFATHER1 | HHX | WTIA | WTFA | INTV_MON | ... | MAFLG | CHFLG | OPFLG | OGFLG | WHONAM1 | WHONAM2 | NOTCOV | PRPLPLUS | PWRKBR1 | COVER | COVER65O | COVER65 | REGIONBR | WHYNOWKP | GEOBRTH | YRSINUS | CITIZENP | DOINGLWP | WRKLYR1 | WRKHRS2 | PLBORN | HEADST | HEADSTV1 | ARMFVER | ARMFEV | ARMFFC | VACOV | WRKFTALL | WRKMYR | HIEMPOF | EDUC1 | ERNYR_P | ARMFTM7P | ARMFTM1P | ARMFTM2P | ARMFTM3P | ARMFTM4P | ARMFTM5P | ARMFTM6P | ENGLANG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAHCA7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | 1.517738 | NaN | 2.355867 | 1.555073 | 1.997081 | 1.592659 | 1.426506 | 2.795367 | 1.674699 | 1.318035 | NaN | 1.011039 | 2.217068 | 2.044158 | 1.897792 | 11.080005 | 1.285413 | 1.0 | NaN | 3.700323 | 1.678244 | 1.426197 | 2.160010 | 2.153560 | 60.266807 | 20.0 | 2018.0 | 3.789878 | 1.200000 | 1.083106 | 1.389831 | 3.318903 | NaN | NaN | 0.148102 | 0.080005 | 27584.910816 | 3896.152890 | 4232.544406 | 6.051848 | ... | 1.0 | NaN | 1.0 | NaN | 1.687939 | 1.369748 | 1.978789 | 2.236436 | 2.617251 | 1.980013 | 2.181214 | 2.602106 | 1.586331 | 5.675154 | 1.242248 | 4.821792 | 1.050112 | 4.173034 | 1.767055 | 34.847765 | 1.129744 | NaN | NaN | 1.307692 | 1.865666 | 1.494343 | 1.653563 | 1.896783 | 10.732941 | 1.566991 | 15.417514 | 19.963181 | 1.77095 | 1.991370 | 1.995331 | 1.860845 | 1.566125 | 1.757522 | 1.723906 | 1.267924 |
| 1 | 1.473255 | NaN | 2.327289 | 1.480508 | 1.997585 | 1.614035 | 1.217391 | 2.777778 | 1.608696 | 1.330916 | NaN | 1.006346 | 2.078876 | 2.015413 | 1.919311 | 11.270172 | 1.317316 | NaN | NaN | 3.475068 | 1.750680 | 1.500453 | 2.216682 | 2.204896 | 67.620127 | 20.0 | 2018.0 | 3.889393 | 1.214286 | 1.160000 | 1.500000 | 3.293744 | NaN | NaN | 0.080689 | 0.040798 | 27113.267452 | 3864.845875 | 4071.953762 | 5.943790 | ... | 1.0 | NaN | 1.0 | NaN | 1.572816 | 1.352941 | 1.970988 | 2.175309 | 1.909091 | 2.099034 | 2.168360 | 2.589260 | 1.435177 | 5.378323 | 1.213055 | 4.900826 | 1.030825 | 4.556664 | 1.870354 | 34.022556 | 1.109701 | NaN | NaN | 1.000000 | 1.786038 | 1.491525 | 1.654762 | 1.884058 | 12.914110 | 1.816667 | 14.697189 | 18.417178 | 1.52500 | 2.117647 | 2.043478 | 1.846809 | 1.581731 | 1.719178 | 1.584416 | 1.261106 |
2 rows × 598 columns
persons_data[["LAHCA7", "LAHCA8"]].describe()
| LAHCA7 | LAHCA8 | |
|---|---|---|
| count | 9288.000000 | 9288.000000 |
| mean | 1.960702 | 2.026055 |
| std | 0.778096 | 0.735485 |
| min | 1.000000 | 1.000000 |
| 25% | 2.000000 | 2.000000 |
| 50% | 2.000000 | 2.000000 |
| 75% | 2.000000 | 2.000000 |
| max | 9.000000 | 9.000000 |
Value counts
persons_data["LAHCA7"].value_counts()
persons_data["LAHCA8"].value_counts()
2 8669
1 496
7 53
9 53
8 17
Name: LAHCA8, dtype: int64
Marital status for heart diseases
Marital Status:
1 - Married - spouse in household
2 - Married - spouse not in household
3 - Married - spouse in household unknown
4 - Widowed
5 - Divorced
6 - Separated
7 - Never married
8 - Living with partner
9 - Unknown marital status
pd.crosstab(persons_data_sub.R_MARITL,persons_data_sub.LAHCA7).plot(kind='bar')
plt.title('Marital status for heart diseases')
plt.xlabel('Heart Disease')
plt.ylabel('Frequency of Marital Status')
Text(0, 0.5, 'Frequency of Marital Status')
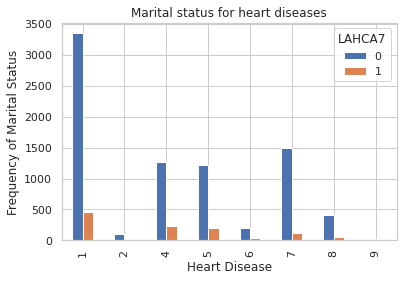
Marital data by different disease groups
pd.crosstab(persons_data.LAHCA7, persons_data.R_MARITL).apply(lambda r: r/r.sum(), axis=1)
| R_MARITL | 1 | 2 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|
| LAHCA7 | ||||||||
| 1 | 0.413418 | 0.009973 | 0.215775 | 0.179510 | 0.031732 | 0.106981 | 0.041704 | 0.000907 |
| 2 | 0.415654 | 0.012404 | 0.157901 | 0.150459 | 0.025056 | 0.184818 | 0.051476 | 0.002233 |
| 7 | 0.471698 | 0.018868 | 0.094340 | 0.132075 | 0.018868 | 0.150943 | 0.075472 | 0.037736 |
| 8 | 0.529412 | 0.058824 | 0.176471 | 0.000000 | 0.000000 | 0.176471 | 0.058824 | 0.000000 |
| 9 | 0.377358 | 0.018868 | 0.075472 | 0.132075 | 0.075472 | 0.188679 | 0.132075 | 0.000000 |
Alcohol abuse data by different disease groups
pd.crosstab(persons_data.LAHCA7, persons_data.LAHCA29_).apply(lambda r: r/r.sum(), axis=1)
| LAHCA29_ | 1 | 2 | 7 | 8 | 9 |
|---|---|---|---|---|---|
| LAHCA7 | |||||
| 1 | 0.000000 | 1.000000 | 0.0 | 0.0 | 0.0 |
| 2 | 0.002109 | 0.997891 | 0.0 | 0.0 | 0.0 |
| 7 | 0.000000 | 0.000000 | 1.0 | 0.0 | 0.0 |
| 8 | 0.000000 | 0.000000 | 0.0 | 1.0 | 0.0 |
| 9 | 0.000000 | 0.000000 | 0.0 | 0.0 | 1.0 |
Health insurance data by different disease groups <65
Under 65 years old
1 - Private
2 - Medicaid and other public
3 - Other coverage
4 - Uninsured
5 - Don’t know
pd.crosstab(persons_data.LAHCA7, persons_data.COVER).apply(lambda r: r/r.sum(), axis=1)
| COVER | 1.0 | 2.0 | 3.0 | 4.0 | 5.0 |
|---|---|---|---|---|---|
| LAHCA7 | |||||
| 1 | 0.301932 | 0.391304 | 0.214976 | 0.089372 | 0.002415 |
| 2 | 0.396811 | 0.338873 | 0.159668 | 0.096789 | 0.007860 |
| 7 | 0.437500 | 0.281250 | 0.156250 | 0.031250 | 0.093750 |
| 8 | 0.750000 | 0.166667 | 0.083333 | 0.000000 | 0.000000 |
| 9 | 0.076923 | 0.538462 | 0.153846 | 0.153846 | 0.076923 |
Health insurance data by different disease groups >65
65 years old and over
1 - Private
2 - Medicaid and other public
3 - Other coverage
4 - Uninsured
5 - Don’t know
pd.crosstab(persons_data.LAHCA7, persons_data.COVER65).apply(lambda r: r/r.sum(), axis=1)
| COVER65 | 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 |
|---|---|---|---|---|---|---|---|
| LAHCA7 | |||||||
| 1 | 0.359942 | 0.142235 | 0.185776 | 0.175617 | 0.133527 | 0.002903 | 0.000000 |
| 2 | 0.352452 | 0.126905 | 0.226378 | 0.164589 | 0.121917 | 0.005542 | 0.002217 |
| 7 | 0.523810 | 0.047619 | 0.000000 | 0.238095 | 0.095238 | 0.000000 | 0.095238 |
| 8 | 0.200000 | 0.000000 | 0.200000 | 0.400000 | 0.200000 | 0.000000 | 0.000000 |
| 9 | 0.407407 | 0.037037 | 0.148148 | 0.185185 | 0.185185 | 0.000000 | 0.037037 |
Private health insurance data by different disease groups
Private health insurance recode
1 - Yes, information
2 - Yes, but no information
3 - No
7 - Refused
8 - Not ascertained
9 - Don’t know
pd.crosstab(persons_data.LAHCA7, persons_data.HIKINDNA).apply(lambda r: r/r.sum(), axis=1)
| HIKINDNA | 1 | 2 | 7 | 9 |
|---|---|---|---|---|
| LAHCA7 | ||||
| 1 | 0.328196 | 0.670898 | 0.000907 | 0.000000 |
| 2 | 0.374969 | 0.619449 | 0.001364 | 0.004217 |
| 7 | 0.509434 | 0.377358 | 0.075472 | 0.037736 |
| 8 | 0.588235 | 0.411765 | 0.000000 | 0.000000 |
| 9 | 0.264151 | 0.679245 | 0.000000 | 0.056604 |
Born in the US data by different disease groups
Born in the United States
1 - Yes
2 - No
7 - Refused
8 - Not ascertained
9 - Don’t know
pd.crosstab(persons_data.LAHCA7, persons_data.PLBORN).apply(lambda r: r/r.sum(), axis=1)
| PLBORN | 1 | 2 | 7 | 9 |
|---|---|---|---|---|
| LAHCA7 | ||||
| 1 | 0.890299 | 0.109701 | 0.000000 | 0.00000 |
| 2 | 0.877078 | 0.121806 | 0.000496 | 0.00062 |
| 7 | 0.943396 | 0.037736 | 0.018868 | 0.00000 |
| 8 | 0.941176 | 0.058824 | 0.000000 | 0.00000 |
| 9 | 0.773585 | 0.226415 | 0.000000 | 0.00000 |
Education data by different disease groups
Highest education level
00-14 - High School and below
15-18 - College
19-21 - Graduate and Above
pd.crosstab(persons_data.LAHCA7, persons_data.EDUC1).apply(lambda r: r/r.sum(), axis=1)
| EDUC1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 97 | 99 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAHCA7 | ||||||||||||||||||||||||
| 1 | 0.004533 | 0.002720 | 0.001813 | 0.006346 | 0.007253 | 0.00544 | 0.014506 | 0.008160 | 0.033545 | 0.033545 | 0.032638 | 0.038078 | 0.028105 | 0.050771 | 0.265639 | 0.198549 | 0.073436 | 0.034451 | 0.093382 | 0.043518 | 0.009066 | 0.005440 | 0.000000 | 0.009066 |
| 2 | 0.008683 | 0.001116 | 0.002853 | 0.004341 | 0.003845 | 0.00707 | 0.012280 | 0.007938 | 0.022947 | 0.024312 | 0.030265 | 0.038452 | 0.027164 | 0.043165 | 0.269784 | 0.180848 | 0.076284 | 0.037212 | 0.121558 | 0.046763 | 0.008559 | 0.010419 | 0.003349 | 0.010791 |
| 7 | 0.018868 | 0.000000 | 0.000000 | 0.018868 | 0.000000 | 0.00000 | 0.000000 | 0.000000 | 0.000000 | 0.018868 | 0.056604 | 0.000000 | 0.018868 | 0.000000 | 0.226415 | 0.169811 | 0.094340 | 0.037736 | 0.113208 | 0.113208 | 0.000000 | 0.000000 | 0.056604 | 0.056604 |
| 8 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.00000 | 0.058824 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.117647 | 0.176471 | 0.058824 | 0.117647 | 0.058824 | 0.411765 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 9 | 0.018868 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.00000 | 0.000000 | 0.000000 | 0.018868 | 0.037736 | 0.037736 | 0.037736 | 0.037736 | 0.056604 | 0.283019 | 0.207547 | 0.056604 | 0.000000 | 0.037736 | 0.075472 | 0.037736 | 0.000000 | 0.018868 | 0.037736 |
Full-time working data by different disease groups
Usually work full time
1 - Yes
2 - No
7 - Refused
8 - Not ascertained
9 - Don’t know
pd.crosstab(persons_data.LAHCA7, persons_data.WRKFTALL).apply(lambda r: r/r.sum(), axis=1)
| WRKFTALL | 1.0 | 2.0 | 9.0 |
|---|---|---|---|
| LAHCA7 | |||
| 1 | 0.115942 | 0.884058 | 0.000000 |
| 2 | 0.131367 | 0.864611 | 0.004021 |
| 7 | 0.000000 | 1.000000 | 0.000000 |
| 8 | 0.666667 | 0.333333 | 0.000000 |
| 9 | 0.222222 | 0.777778 | 0.000000 |
Statistical variables of dataset
persons_data_analysis = persons_data_sub[['ENGLANG', 'PSSRR', 'EDUC1',
'CITIZENP','PLBORN', 'HIKINDNA', 'LAHCA29_', 'R_MARITL', 'AGE_P',
'MRACRPI2', 'SEX', 'WTFA','LAHCA7']]
percent_missing = persons_data_analysis.isnull().sum() * 100 / len(persons_data_analysis)
missing_value_persons_data = pd.DataFrame({'column_name': persons_data_analysis.columns,
'percent_missing': percent_missing})
missing_value_persons_data.sort_values('percent_missing', inplace=True)
print(missing_value_persons_data)
column_name percent_missing
ENGLANG ENGLANG 0.0
PSSRR PSSRR 0.0
EDUC1 EDUC1 0.0
CITIZENP CITIZENP 0.0
PLBORN PLBORN 0.0
HIKINDNA HIKINDNA 0.0
LAHCA29_ LAHCA29_ 0.0
R_MARITL R_MARITL 0.0
AGE_P AGE_P 0.0
MRACRPI2 MRACRPI2 0.0
SEX SEX 0.0
WTFA WTFA 0.0
LAHCA7 LAHCA7 0.0
X = persons_data_analysis.loc[:, persons_data_analysis.columns != 'LAHCA7']
y = persons_data_analysis.loc[:, persons_data_analysis.columns == 'LAHCA7']
import statsmodels.api as sm
logit_model=sm.Logit(y,X)
result=logit_model.fit()
print(result.summary2())
Optimization terminated successfully.
Current function value: 0.354418
Iterations 7
Results: Logit
=================================================================
Model: Logit Pseudo R-squared: 0.036
Dependent Variable: LAHCA7 AIC: 6520.4878
Date: 2020-08-09 16:10 BIC: 6605.9656
No. Observations: 9165 Log-Likelihood: -3248.2
Df Model: 11 LL-Null: -3369.2
Df Residuals: 9153 LLR p-value: 1.3268e-45
Converged: 1.0000 Scale: 1.0000
No. Iterations: 7.0000
------------------------------------------------------------------
Coef. Std.Err. z P>|z| [0.025 0.975]
------------------------------------------------------------------
ENGLANG 0.0202 0.0474 0.4267 0.6696 -0.0727 0.1132
PSSRR -0.0723 0.0499 -1.4491 0.1473 -0.1701 0.0255
EDUC1 -0.0090 0.0040 -2.2592 0.0239 -0.0168 -0.0012
CITIZENP -0.0913 0.1415 -0.6456 0.5185 -0.3687 0.1860
PLBORN -0.2753 0.1290 -2.1336 0.0329 -0.5281 -0.0224
HIKINDNA 0.0629 0.0490 1.2850 0.1988 -0.0330 0.1589
LAHCA29_ -1.3984 0.1428 -9.7949 0.0000 -1.6782 -1.1185
R_MARITL 0.0184 0.0143 1.2878 0.1978 -0.0096 0.0464
AGE_P 0.0290 0.0024 11.9857 0.0000 0.0243 0.0338
MRACRPI2 0.0306 0.0135 2.2579 0.0240 0.0040 0.0572
SEX -0.3917 0.0658 -5.9491 0.0000 -0.5207 -0.2627
WTFA -0.0000 0.0000 -0.8577 0.3911 -0.0001 0.0000
=================================================================
Logistic regression
from sklearn.linear_model import LogisticRegression
from sklearn import metrics
X_train, X_test, y_train, y_test = train_test_split(X, y.values.ravel(), test_size=0.1, random_state=0)
logreg = LogisticRegression(solver='lbfgs', max_iter=1000)
logreg.fit(X_train, y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=1000,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False)
y_pred = logreg.predict(X_test)
print('Accuracy of logistic regression classifier on test set: {:.2f}'.format(logreg.score(X_test, y_test)))
Accuracy of logistic regression classifier on test set: 0.89
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
precision recall f1-score support
0 0.89 1.00 0.94 817
1 0.00 0.00 0.00 100
accuracy 0.89 917
macro avg 0.45 0.50 0.47 917
weighted avg 0.79 0.89 0.84 917
/usr/local/lib/python3.6/dist-packages/sklearn/metrics/_classification.py:1272: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
Calculated mean by different groups and age
LAHCA7: Persons 18+ years who have at least one limitation due to a heart problem
Heart problem causes limitation:
1 - Mentioned
2 - Not mentioned
7 - Refused
8 - Not ascertained
9 - Don’t know
persons_data.groupby('LAHCA7')['AGE_P'].mean()
LAHCA7
1 67.620127
2 60.266807
7 57.905660
8 59.705882
9 62.056604
Name: AGE_P, dtype: float64
Calculated mean by different incomes
Total earnings last year
01 - $01-$4,999
02 - $5,000-$9,999
03 - $10,000-$14,999
04 - $15,000-$19,999
05 - $20,000-$24,999
06 - $25,000-$34,999
07 - $35,000-$44,999
08 - $45,000-$54,999
09 - $55,000-$64,999
10 - $65,000-$74,999
11 - $75,000 and over
97 - Refused
98 - Not ascertained
99 - Don’t know
persons_data.groupby('LAHCA7')['ERNYR_P'].median()
LAHCA7
1 6.0
2 6.0
7 97.0
8 8.0
9 6.0
Name: ERNYR_P, dtype: float64
Calculated mean by different language levels
How well English is spoken
1 - Very well
2 - Well
3 - Not well
4 - Not at all
7 - Refused
8- Not ascertained
9 - Don’t know
persons_data.groupby('LAHCA7')['ENGLANG'].median()
LAHCA7
1 1
2 1
7 1
8 1
9 1
Name: ENGLANG, dtype: int64
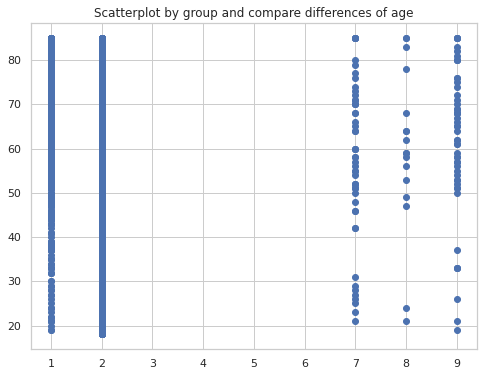
Scatterplot by group and compare differences of age
fig, ax = plt.subplots(figsize=(8,6))
ax.scatter(persons_data.LAHCA7, persons_data.AGE_P, cmap='tab20b')
plt.title('Scatterplot by group and compare differences of age')
plt.show()
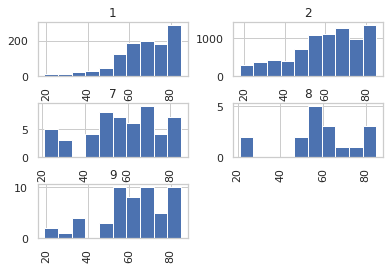
Histogram of Ages
persons_data['AGE_P'].hist(by=persons_data['LAHCA7'])
array([[<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3c65850f0>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3d3e8ee48>],
[<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3d3ed4908>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3c6220390>],
[<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3c678da90>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7fc3c617f588>]],
dtype=object)
About
Project by Max Shalom
Source code and data available on the GitHub Repository
| Home | Notebook | Data Table | Poster |